View 2D Alignment
This menu is used for secondary structure visualization whereby you input your aligned sequences and secondary structure predictions. You can either insert your data directly in the text boxes or upload files for each input data. If all fields are filled, only your uploaded files will be considered.
Data format for text boxes
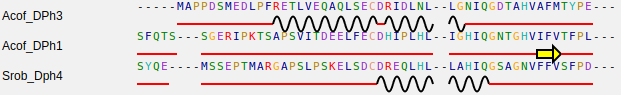
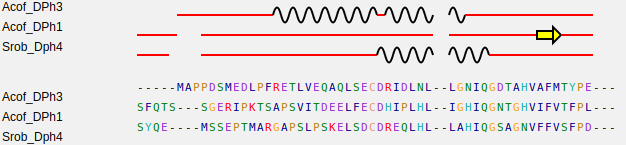
For the sequence text box, each line must contain the protein name followed by spaces followed by the amino acids sequence. See an example bellow.
.For the 2D Structures text box, each line must contain the protein name followed by a tabulation followed by the secondary structure prediction. See an example bellow, where C stand for "loops", H for alpha-helixes and E for beta-sheets.
Make sure the protein names correspond to those inserted in the sequence text box, and the length of secondary structure prediction is the same of amino acid sequence without gaps. Sequences in the two boxes could not appear in the same order. Note that the secondary structures may only contain C,H and E.
For other examples, please see the Examples tab.
Uploading files
Alternatively, you may upload the alignment and 2D Structure files instead of filling boxes. For that, click on the appropriate upload buttons, see Figure below.
The uploaded files must respect the same format of the text boxes, see above.
Alignment 2D File
The third option available is to upload the alignment 2D file generated from the Ali2D website https://toolkit.tuebingen.mpg.de/#/tools/ali2d. In the Ali2D website, you can insert your unaligned sequences and the tool will align them and predict their 2D structures. Our website accept their output file and produce an appropriate visualization. To obtain Ali2D output, click on the tab "Text Output" and save all information in a local file. Next, the local file can be uploaded on our website, see below:
Additional parameters
You may choose the output size you wish for the result.
You may also wish to separate the sequences block input from the visual representation on the result page. Check the separate box to do so.
Submitting
Once you have ensured all your data or files have been correctly input, click on the button and the result will appear on a new page.