:: Input ::
:: Output ::
- List of genes
- Protein architecture
- Explore architectures in Plasmobase
- Explore architectures in Uniprot
- Compare architectures in Plasmobase
- Compare architectures in Uniprot
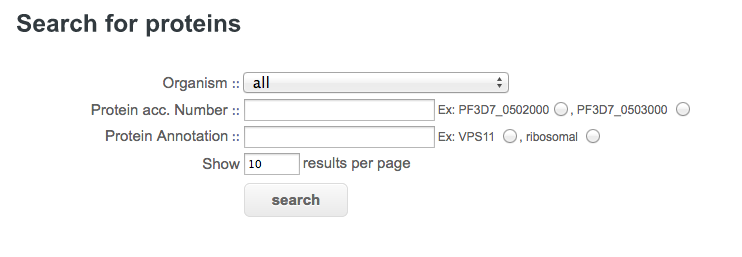
Domain architectures are accessed by providing a protein/gene identifier. The list of descriptors is given below.
|
Organism : select a Plasmodium species or all of them for a search on a single genome or on all fully sequenced genomes of PlasmoDB.
Protein acc. Number : enter a protein/gene accession number of PlasmoDB. For instance, PF3D7_0906400 or click on the radio button PF3D7_0502000 or PF3D7_0503000 for automatic selection.
Protein Annotation : enter a protein/gene annotation descriptor of PlasmoDB. For instance, words like "cyclin", "kinase binding protein", "unknown function" or click on the radio button VPS11 or ribosomal for automatic selection.
Show results per page : number of results displayed per page.
|
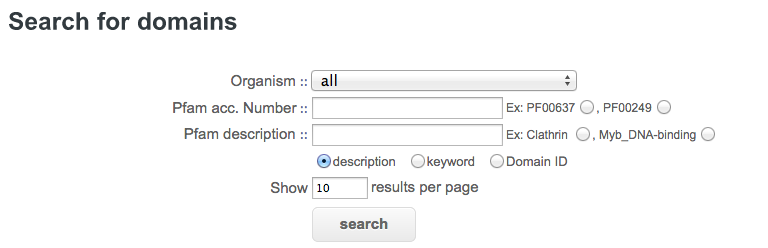
Organism : select a Plasmodium species or all of them for a search on a single genome or on all fully sequenced genomes of PlasmoDB.
Pfam acc. Number : enter a Pfam accession number. For instance: PF08613 or click on the radio button PF00637 or PF00249 for automatic selection.
Pfam description : enter either a Pfam descriptor, a keyword or a domain id. For instance: DEAD or click on the radio button Clathrin or Myb_DNA-binding for automatic selection. You can specify the Pfam text field for your search, “Pfam description” searches in all text data in the Pfam database, “keyword” searches only in keyword fields, and “Domain Id” searches only in domain name fields.
Show results per page : number of results displayed per page.
|
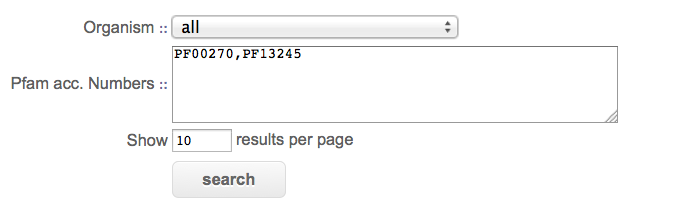
Organism : select a Plasmodium species or all of them for a search on a single genome or on all fully sequenced genomes of PlasmoDB.
Pfam acc. Number : enter at least two Pfam accession numbers. For instance, PF00270,PF13245. Accession numbers are separated by a comma without spaces nor breaklines.
Show results per page : number of results displayed per page.
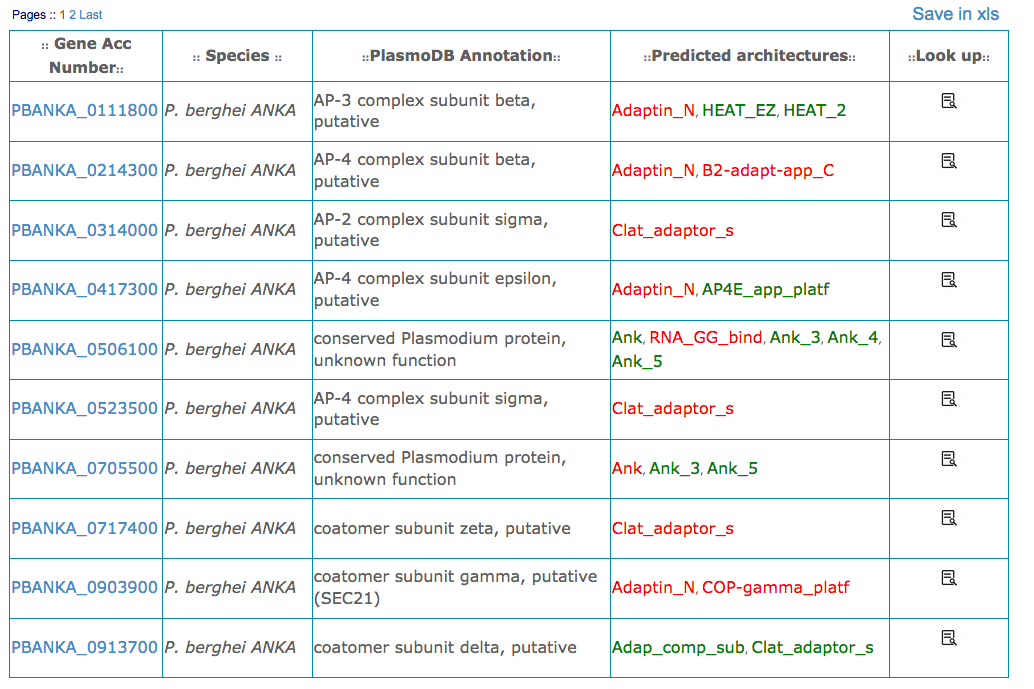
The search (by proteins, domains or domain architectures) provides a list of genes and their domain architectures. For exploring the architectures, click icons in the "Look up" column. For save them in a xls format click on "Save in xls".
|
Gene Acc Number : genes accession number according to PlasmoDB. Active link to the PlasmoDB website.
Species : Plasmodium organism name
PlasmoDB Annotation : gene annotation provided by PlasmoDB
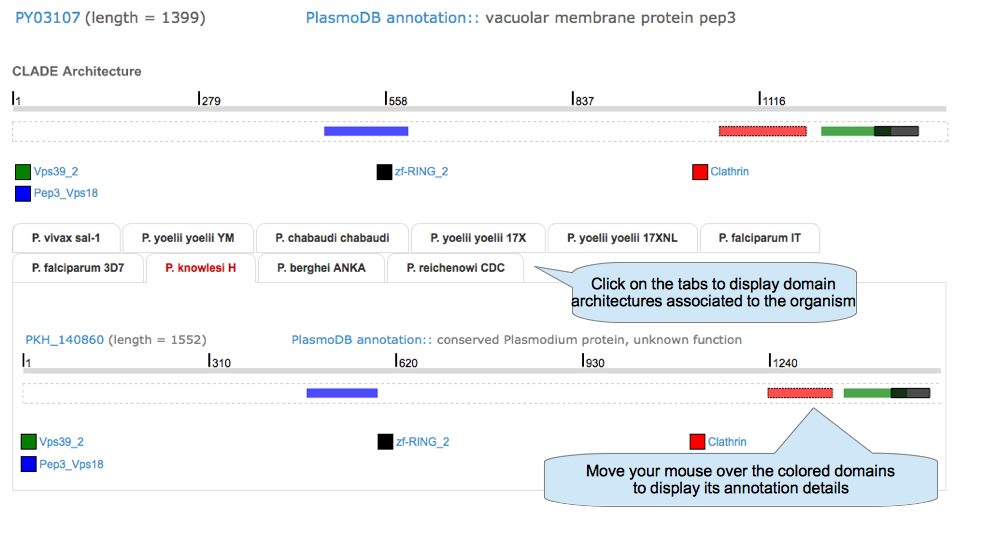
Predicted architectures : CLADE domain architecture. Domains found also by Pfam are in red, new domains detected by CLADE are in green. Active link to the Pfam website.
Look up : Access to the protein architecture page
Show details for domain architecture of a given protein.
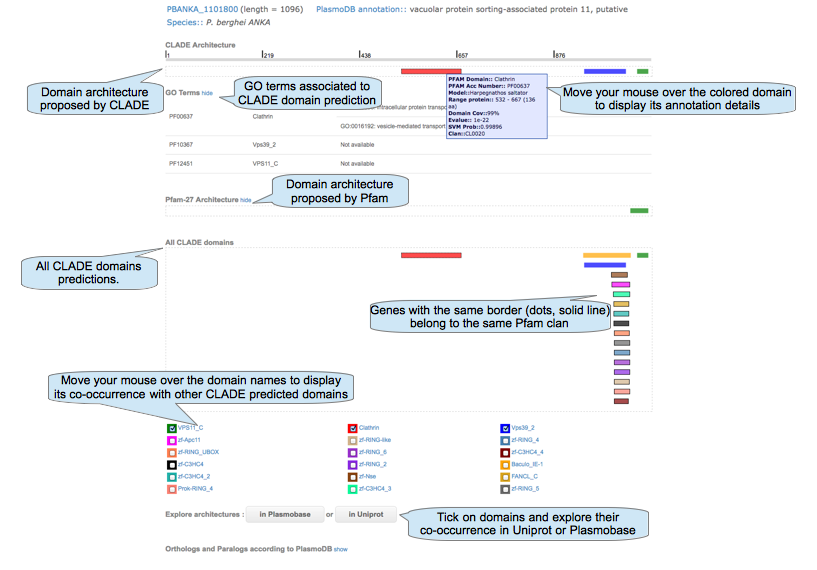
|
We show also a list of orthologs and paralogs according to PlasmoDB. The gene accession number is linked to its protein architecture page.
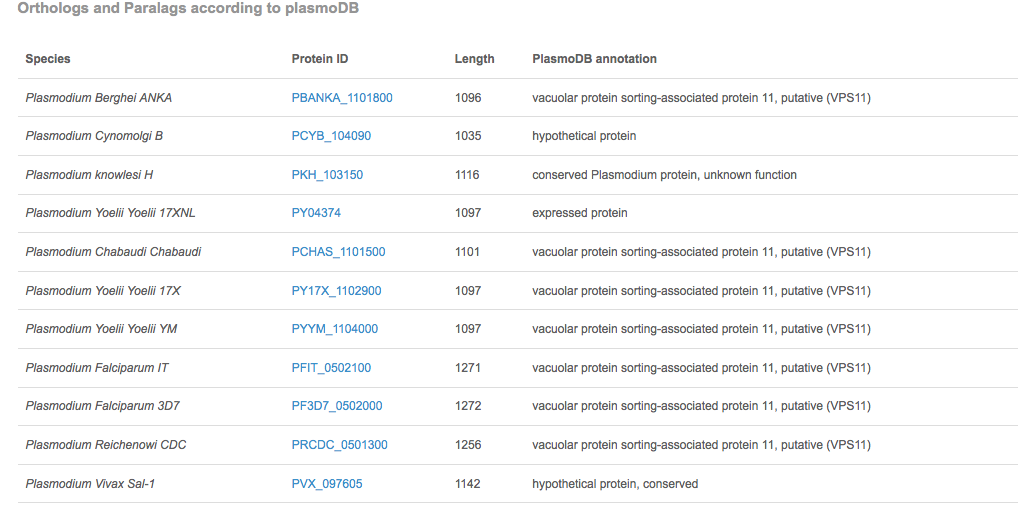
|
After selecting some domains in the protein architecture page and clicking on the button "In Plasmobase", you can explore the occurrence of that architecture in all Plasmodium organisms and compare them.
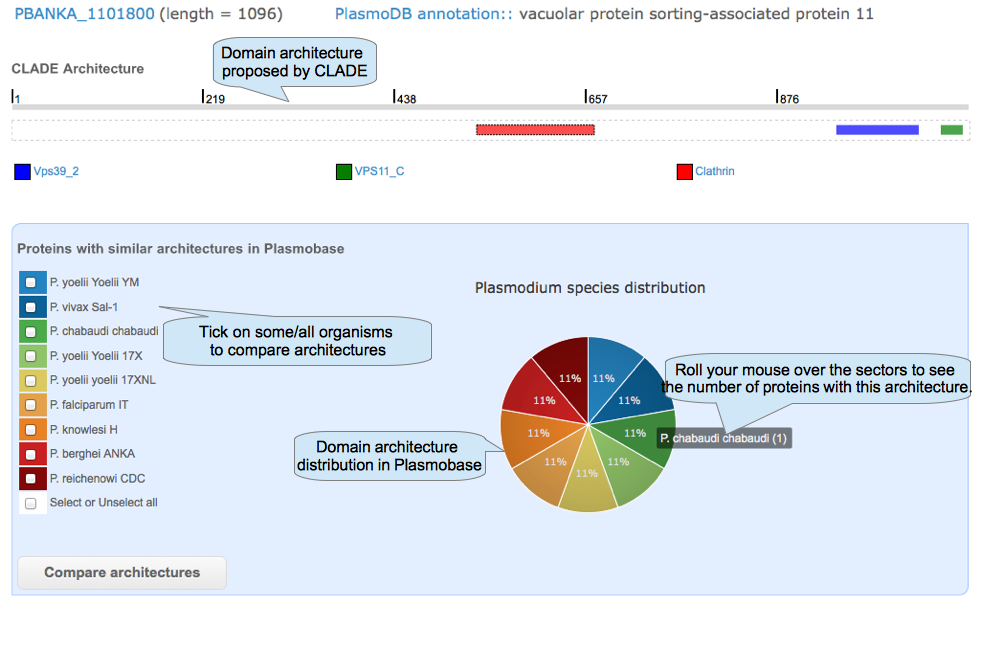
|
After selecting some domains in the protein architecture page and clicking on the button "In Uniprot", you can explore the occurrence of that architecture in all species available in Uniprot to compare them.
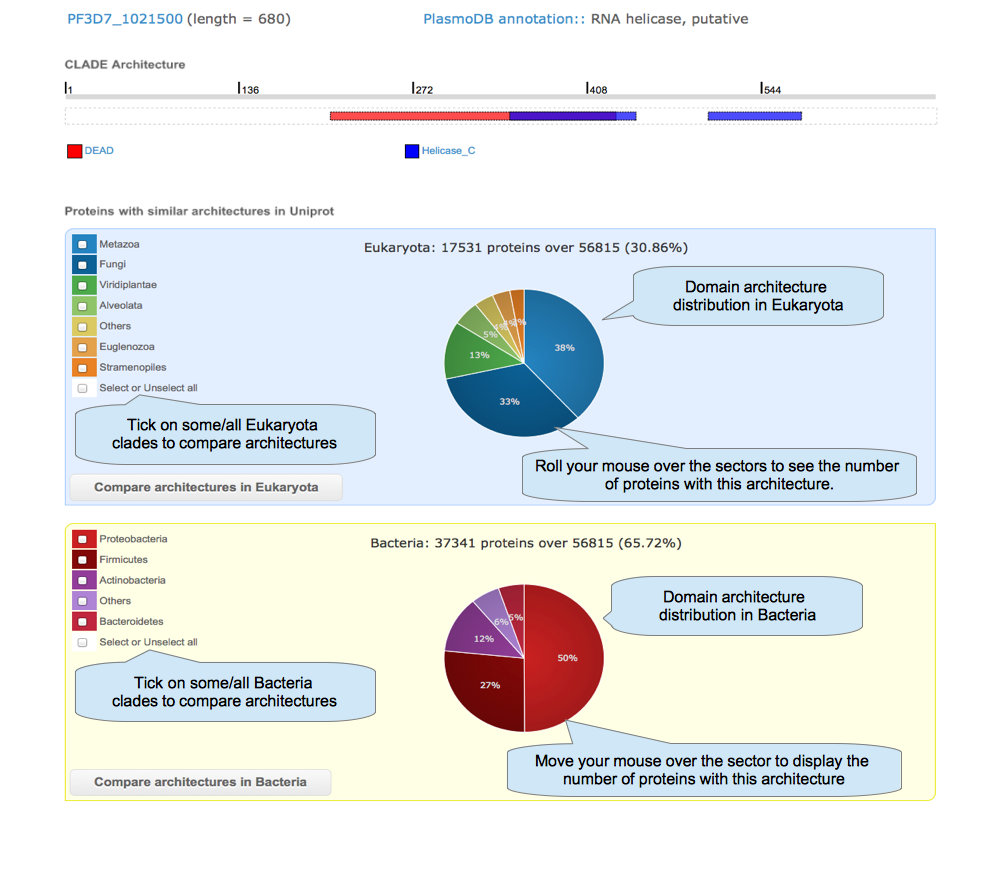
|
After selecting some/all species in the "Explore architectures in Plasmobase" page and clicking on the button "Compare architectures", you can visualize domain architectures for the selected species and compare them to the query sequence architecture.
|
